Explore Workflows
View already parsed workflows here or click here to add your own
| Graph | Name | Retrieved From | View |
|---|---|---|---|
|
|
QuantSeq 3' mRNA-Seq single-read
### Pipeline for Lexogen's QuantSeq 3' mRNA-Seq Library Prep Kit FWD for Illumina [Lexogen original documentation](https://www.lexogen.com/quantseq-3mrna-sequencing/) * Cost-saving and streamlined globin mRNA depletion during QuantSeq library preparation * Genome-wide analysis of gene expression * Cost-efficient alternative to microarrays and standard RNA-Seq * Down to 100 pg total RNA input * Applicable for low quality and FFPE samples * Single-read sequencing of up to 9,216 samples/lane * Dual indexing and Unique Molecular Identifiers (UMIs) are available ### QuantSeq 3’ mRNA-Seq Library Prep Kit FWD for Illumina The QuantSeq FWD Kit is a library preparation protocol designed to generate Illumina compatible libraries of sequences close to the 3’ end of polyadenylated RNA. QuantSeq FWD contains the Illumina Read 1 linker sequence in the second strand synthesis primer, hence NGS reads are generated towards the poly(A) tail, directly reflecting the mRNA sequence (see workflow). This version is the recommended standard for gene expression analysis. Lexogen furthermore provides a high-throughput version with optional dual indexing (i5 and i7 indices) allowing up to 9,216 samples to be multiplexed in one lane. #### Analysis of Low Input and Low Quality Samples The required input amount of total RNA is as low as 100 pg. QuantSeq is suitable to reproducibly generate libraries from low quality RNA, including FFPE samples. See Fig.1 and 2 for a comparison of two different RNA qualities (FFPE and fresh frozen cryo-block) of the same sample. 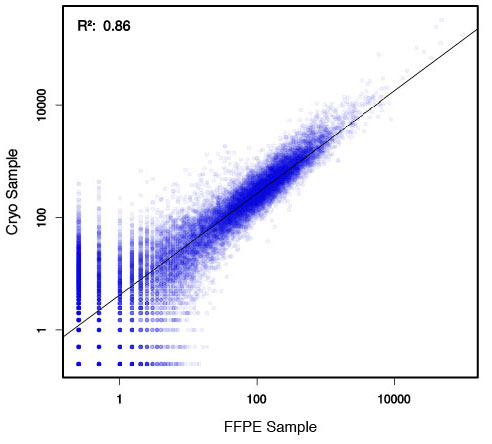 Figure 1 | Correlation of gene counts of FFPE and cryo samples. 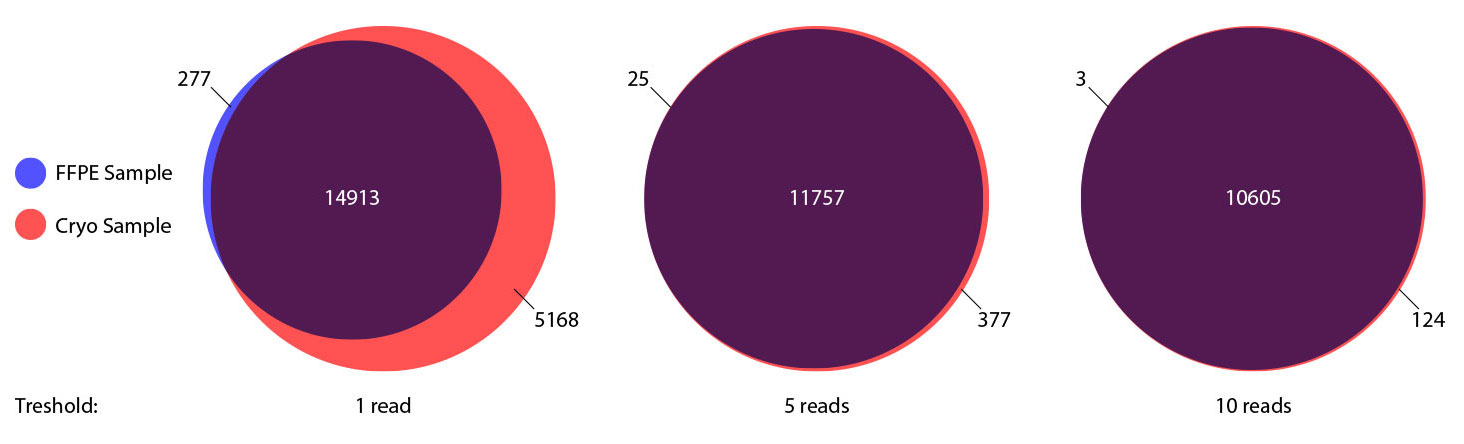 Figure 2 | Venn diagrams of genes detected by QuantSeq at a uniform read depth of 2.5 M reads in FFPE and cryo samples with 1, 5, and 10 reads/gene thresholds. #### Mapping of Transcript End Sites By using longer reads QuantSeq FWD allows to exactly pinpoint the 3’ end of poly(A) RNA (see Fig. 3) and therefore obtain accurate information about the 3’ UTR. 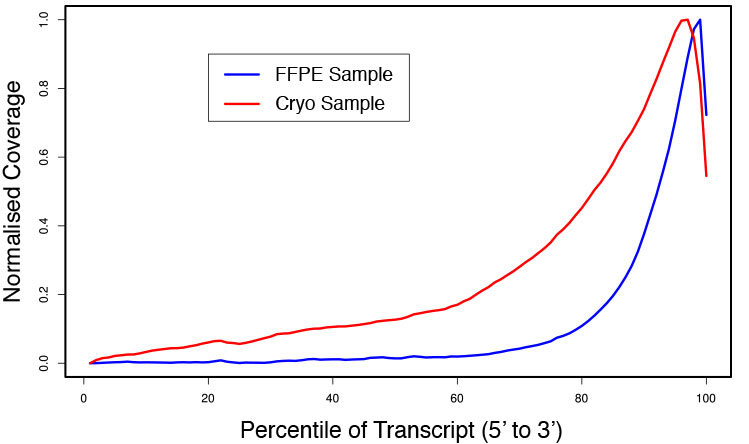 Figure 3 | QuantSeq read coverage versus normalized transcript length of NGS libraries derived from FFPE-RNA (blue) and cryo-preserved RNA (red). ### Current workflow should be used only with the single-end RNA-Seq data. It performs the following steps: 1. Separates UMIes and trims adapters from input FASTQ file 2. Uses ```STAR``` to align reads from input FASTQ file according to the predefined reference indices; generates unsorted BAM file and alignment statistics file 3. Uses ```fastx_quality_stats``` to analyze input FASTQ file and generates quality statistics file 4. Uses ```samtools sort``` and generates coordinate sorted BAM(+BAI) file pair from the unsorted BAM file obtained on the step 2 (after running STAR) 5. Uses ```umi_tools dedup``` and generates final filtered sorted BAM(+BAI) file pair 6. Generates BigWig file on the base of sorted BAM file 7. Maps input FASTQ file to predefined rRNA reference indices using ```bowtie``` to define the level of rRNA contamination; exports resulted statistics to file 8. Calculates isoform expression level for the sorted BAM file and GTF/TAB annotation file using GEEP reads-counting utility; exports results to file |
Path: workflows/trim-quantseq-mrnaseq-se.cwl Branch/Commit ID: 2005c6b7f1bff6247d015ff6c116bd9ec97158bb |
|
|
|
RNA-Seq pipeline paired-end
The original [BioWardrobe's](https://biowardrobe.com) [PubMed ID:26248465](https://www.ncbi.nlm.nih.gov/pubmed/26248465) **RNA-Seq** basic analysis for a **paired-end** experiment. A corresponded input [FASTQ](http://maq.sourceforge.net/fastq.shtml) file has to be provided. Current workflow should be used only with the paired-end RNA-Seq data. It performs the following steps: 1. Use STAR to align reads from input FASTQ files according to the predefined reference indices; generate unsorted BAM file and alignment statistics file 2. Use fastx_quality_stats to analyze input FASTQ files and generate quality statistics files 3. Use samtools sort to generate coordinate sorted BAM(+BAI) file pair from the unsorted BAM file obtained on the step 1 (after running STAR) 4. Generate BigWig file on the base of sorted BAM file 5. Map input FASTQ files to predefined rRNA reference indices using Bowtie to define the level of rRNA contamination; export resulted statistics to file 6. Calculate isoform expression level for the sorted BAM file and GTF/TAB annotation file using GEEP reads-counting utility; export results to file |
Path: workflows/rnaseq-pe.cwl Branch/Commit ID: 2005c6b7f1bff6247d015ff6c116bd9ec97158bb |
|
|
|
Build STAR indices
Workflow runs [STAR](https://github.com/alexdobin/STAR) v2.5.3a (03/17/2017) PMID: [23104886](https://www.ncbi.nlm.nih.gov/pubmed/23104886) to build indices for reference genome provided in a single FASTA file as fasta_file input and GTF annotation file from annotation_gtf_file input. Generated indices are saved in a folder with the name that corresponds to the input genome. |
Path: workflows/star-index.cwl Branch/Commit ID: 2005c6b7f1bff6247d015ff6c116bd9ec97158bb |
|
|
|
SoupX Estimate
SoupX Estimate ============== |
Path: workflows/soupx.cwl Branch/Commit ID: 2005c6b7f1bff6247d015ff6c116bd9ec97158bb |
|
|
|
THOR - differential peak calling of ChIP-seq signals with replicates
What is THOR? -------------- THOR is an HMM-based approach to detect and analyze differential peaks in two sets of ChIP-seq data from distinct biological conditions with replicates. THOR performs genomic signal processing, peak calling and p-value calculation in an integrated framework. For more information please refer to: ------------------------------------- Allhoff, M., Sere K., Freitas, J., Zenke, M., Costa, I.G. (2016), Differential Peak Calling of ChIP-seq Signals with Replicates with THOR, Nucleic Acids Research, epub gkw680. |
Path: workflows/rgt-thor.cwl Branch/Commit ID: 2005c6b7f1bff6247d015ff6c116bd9ec97158bb |
|
|
|
Genomic regions intersection and visualization
Genomic regions intersection and visualization ============================================== 1. Merges intervals within each of the filtered peaks files from ChIP/ATAC experiments 2. Overlaps merged intervals and assigns the nearest genes to them |
Path: workflows/intervene.cwl Branch/Commit ID: 2005c6b7f1bff6247d015ff6c116bd9ec97158bb |
|
|
|
ChIP-Seq pipeline paired-end
The original [BioWardrobe's](https://biowardrobe.com) [PubMed ID:26248465](https://www.ncbi.nlm.nih.gov/pubmed/26248465) **ChIP-Seq** basic analysis workflow for a **paired-end** experiment. A [FASTQ](http://maq.sourceforge.net/fastq.shtml) input file has to be provided. The pipeline produces a sorted BAM file alongside with index BAI file, quality statistics of the input FASTQ file, coverage by estimated fragments as a BigWig file, peaks calling data in a form of narrowPeak or broadPeak files, islands with the assigned nearest genes and region type, data for average tag density plot. Workflow starts with step *fastx\_quality\_stats* from FASTX-Toolkit to calculate quality statistics for input FASTQ file. At the same time `bowtie` is used to align reads from input FASTQ file to reference genome *bowtie\_aligner*. The output of this step is an unsorted SAM file which is being sorted and indexed by `samtools sort` and `samtools index` *samtools\_sort\_index*. Depending on workflow’s input parameters indexed and sorted BAM file can be processed by `samtools rmdup` *samtools\_rmdup* to get rid of duplicated reads. If removing duplicates is not required the original BAM and BAI files are returned. Otherwise step *samtools\_sort\_index\_after\_rmdup* repeat `samtools sort` and `samtools index` with BAM and BAI files without duplicates. Next `macs2 callpeak` performs peak calling *macs2\_callpeak* and the next step reports *macs2\_island\_count* the number of islands and estimated fragment size. If the latter is less that 80bp (hardcoded in the workflow) `macs2 callpeak` is rerun again with forced fixed fragment size value (*macs2\_callpeak\_forced*). It is also possible to force MACS2 to use pre set fragment size in the first place. Next step (*macs2\_stat*) is used to define which of the islands and estimated fragment size should be used in workflow output: either from *macs2\_island\_count* step or from *macs2\_island\_count\_forced* step. If input trigger of this step is set to True it means that *macs2\_callpeak\_forced* step was run and it returned different from *macs2\_callpeak* step results, so *macs2\_stat* step should return [fragments\_new, fragments\_old, islands\_new], if trigger is False the step returns [fragments\_old, fragments\_old, islands\_old], where sufix \"old\" defines results obtained from *macs2\_island\_count* step and sufix \"new\" - from *macs2\_island\_count\_forced* step. The following two steps (*bamtools\_stats* and *bam\_to\_bigwig*) are used to calculate coverage from BAM file and save it in BigWig format. For that purpose bamtools stats returns the number of mapped reads which is then used as scaling factor by bedtools genomecov when it performs coverage calculation and saves it as a BEDgraph file whichis then sorted and converted to BigWig format by bedGraphToBigWig tool from UCSC utilities. Step *get\_stat* is used to return a text file with statistics in a form of [TOTAL, ALIGNED, SUPRESSED, USED] reads count. Step *island\_intersect* assigns nearest genes and regions to the islands obtained from *macs2\_callpeak\_forced*. Step *average\_tag\_density* is used to calculate data for average tag density plot from the BAM file. |
Path: workflows/chipseq-pe.cwl Branch/Commit ID: 2005c6b7f1bff6247d015ff6c116bd9ec97158bb |
|
|
|
Motif Finding with HOMER with random background regions
Motif Finding with HOMER with random background regions --------------------------------------------------- HOMER contains a novel motif discovery algorithm that was designed for regulatory element analysis in genomics applications (DNA only, no protein). It is a differential motif discovery algorithm, which means that it takes two sets of sequences and tries to identify the regulatory elements that are specifically enriched in on set relative to the other. It uses ZOOPS scoring (zero or one occurrence per sequence) coupled with the hypergeometric enrichment calculations (or binomial) to determine motif enrichment. HOMER also tries its best to account for sequenced bias in the dataset. It was designed with ChIP-Seq and promoter analysis in mind, but can be applied to pretty much any nucleic acids motif finding problem. Here is how we generate background for Motifs Analysis ------------------------------------- 1. Take input file with regions in a form of “chr\" “start\" “end\" 2. Sort and remove duplicates from this regions file 3. Extend each region in 20Kb into both directions 4. Merge all overlapped extended regions 5. Subtract not extended regions from the extended ones 6. Randomly distribute not extended regions within the regions that we got as a result of the previous step 7. Get fasta file from these randomly distributed regions (from the previous step). Use it as background For more information please refer to: ------------------------------------- [Official documentation](http://homer.ucsd.edu/homer/motif/) |
Path: workflows/homer-motif-analysis.cwl Branch/Commit ID: 2005c6b7f1bff6247d015ff6c116bd9ec97158bb |
|
|
|
DiffBind - Differential Binding Analysis of ChIP-Seq Peak Data
Differential Binding Analysis of ChIP-Seq Peak Data --------------------------------------------------- DiffBind processes ChIP-Seq data enriched for genomic loci where specific protein/DNA binding occurs, including peak sets identified by ChIP-Seq peak callers and aligned sequence read datasets. It is designed to work with multiple peak sets simultaneously, representing different ChIP experiments (antibodies, transcription factor and/or histone marks, experimental conditions, replicates) as well as managing the results of multiple peak callers. For more information please refer to: ------------------------------------- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, Ali S, Chin S, Palmieri C, Caldas C, Carroll JS (2012). “Differential oestrogen receptor binding is associated with clinical outcome in breast cancer.” Nature, 481, -4. |
Path: workflows/diffbind.cwl Branch/Commit ID: 2005c6b7f1bff6247d015ff6c116bd9ec97158bb |
|
|
|
FastQC - a quality control tool for high throughput sequence data
FastQC - a quality control tool for high throughput sequence data ===================================== FastQC aims to provide a simple way to do some quality control checks on raw sequence data coming from high throughput sequencing pipelines. It provides a modular set of analyses which you can use to give a quick impression of whether your data has any problems of which you should be aware before doing any further analysis. The main functions of FastQC are: - Import of data from FastQ files (any variant) - Providing a quick overview to tell you in which areas there may be problems - Summary graphs and tables to quickly assess your data - Export of results to an HTML based permanent report - Offline operation to allow automated generation of reports without running the interactive application |
Path: workflows/fastqc.cwl Branch/Commit ID: 2005c6b7f1bff6247d015ff6c116bd9ec97158bb |




