Explore Workflows
View already parsed workflows here or click here to add your own
| Graph | Name | Retrieved From | View |
|---|---|---|---|
|
|
Feature expression merge - combines feature expression from several experiments
Feature expression merge - combines feature expression from several experiments ========================================================================= Workflows merges RPKM (by default) gene expression from several experiments based on the values from GeneId, Chrom, TxStart, TxEnd and Strand columns (by default). Reported unique columns are renamed based on the experiments names. |
Path: workflows/feature-merge.cwl Branch/Commit ID: 935a78f1aff757f977de4e3672aefead3b23606b |
|
|
|
RNA-Seq pipeline paired-end strand specific
The original [BioWardrobe's](https://biowardrobe.com) [PubMed ID:26248465](https://www.ncbi.nlm.nih.gov/pubmed/26248465) **RNA-Seq** basic analysis for a **paired-end** experiment. A corresponded input [FASTQ](http://maq.sourceforge.net/fastq.shtml) file has to be provided. Current workflow should be used only with the paired-end RNA-Seq data. It performs the following steps: 1. Use STAR to align reads from input FASTQ files according to the predefined reference indices; generate unsorted BAM file and alignment statistics file 2. Use fastx_quality_stats to analyze input FASTQ files and generate quality statistics files 3. Use samtools sort to generate coordinate sorted BAM(+BAI) file pair from the unsorted BAM file obtained on the step 1 (after running STAR) 4. Generate BigWig file on the base of sorted BAM file 5. Map input FASTQ files to predefined rRNA reference indices using Bowtie to define the level of rRNA contamination; export resulted statistics to file 6. Calculate isoform expression level for the sorted BAM file and GTF/TAB annotation file using GEEP reads-counting utility; export results to file |
Path: workflows/rnaseq-pe-dutp.cwl Branch/Commit ID: 935a78f1aff757f977de4e3672aefead3b23606b |
|
|
|
QuantSeq 3' mRNA-Seq single-read
### Pipeline for Lexogen's QuantSeq 3' mRNA-Seq Library Prep Kit FWD for Illumina [Lexogen original documentation](https://www.lexogen.com/quantseq-3mrna-sequencing/) * Cost-saving and streamlined globin mRNA depletion during QuantSeq library preparation * Genome-wide analysis of gene expression * Cost-efficient alternative to microarrays and standard RNA-Seq * Down to 100 pg total RNA input * Applicable for low quality and FFPE samples * Single-read sequencing of up to 9,216 samples/lane * Dual indexing and Unique Molecular Identifiers (UMIs) are available ### QuantSeq 3’ mRNA-Seq Library Prep Kit FWD for Illumina The QuantSeq FWD Kit is a library preparation protocol designed to generate Illumina compatible libraries of sequences close to the 3’ end of polyadenylated RNA. QuantSeq FWD contains the Illumina Read 1 linker sequence in the second strand synthesis primer, hence NGS reads are generated towards the poly(A) tail, directly reflecting the mRNA sequence (see workflow). This version is the recommended standard for gene expression analysis. Lexogen furthermore provides a high-throughput version with optional dual indexing (i5 and i7 indices) allowing up to 9,216 samples to be multiplexed in one lane. #### Analysis of Low Input and Low Quality Samples The required input amount of total RNA is as low as 100 pg. QuantSeq is suitable to reproducibly generate libraries from low quality RNA, including FFPE samples. See Fig.1 and 2 for a comparison of two different RNA qualities (FFPE and fresh frozen cryo-block) of the same sample. 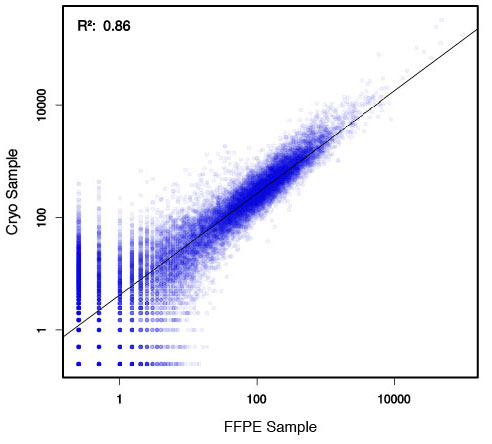 Figure 1 | Correlation of gene counts of FFPE and cryo samples. 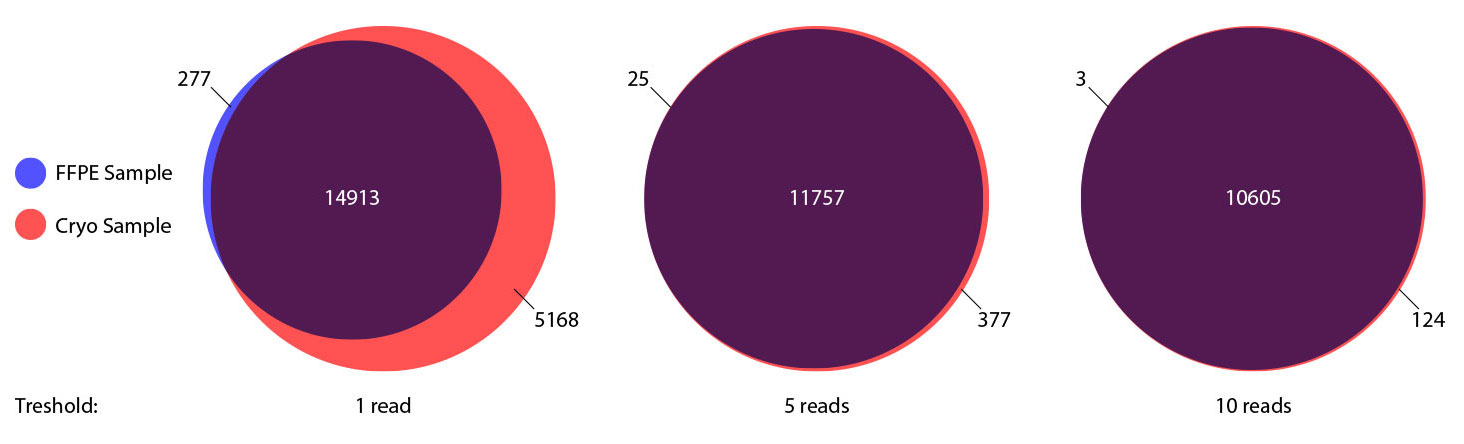 Figure 2 | Venn diagrams of genes detected by QuantSeq at a uniform read depth of 2.5 M reads in FFPE and cryo samples with 1, 5, and 10 reads/gene thresholds. #### Mapping of Transcript End Sites By using longer reads QuantSeq FWD allows to exactly pinpoint the 3’ end of poly(A) RNA (see Fig. 3) and therefore obtain accurate information about the 3’ UTR. 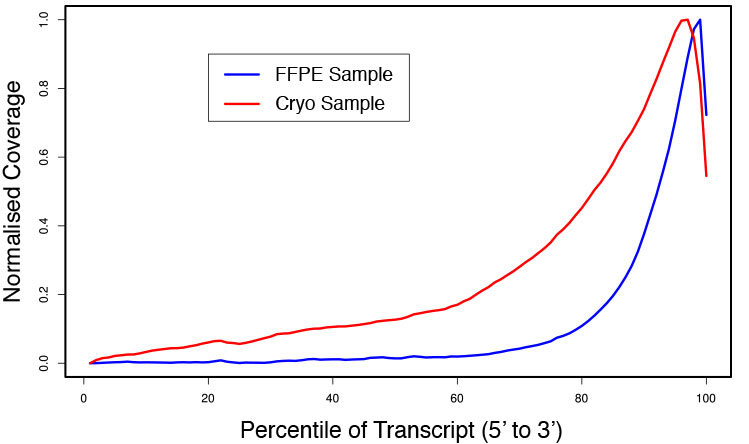 Figure 3 | QuantSeq read coverage versus normalized transcript length of NGS libraries derived from FFPE-RNA (blue) and cryo-preserved RNA (red). ### Current workflow should be used only with the single-end RNA-Seq data. It performs the following steps: 1. Separates UMIes and trims adapters from input FASTQ file 2. Uses ```STAR``` to align reads from input FASTQ file according to the predefined reference indices; generates unsorted BAM file and alignment statistics file 3. Uses ```fastx_quality_stats``` to analyze input FASTQ file and generates quality statistics file 4. Uses ```samtools sort``` and generates coordinate sorted BAM(+BAI) file pair from the unsorted BAM file obtained on the step 2 (after running STAR) 5. Uses ```umi_tools dedup``` and generates final filtered sorted BAM(+BAI) file pair 6. Generates BigWig file on the base of sorted BAM file 7. Maps input FASTQ file to predefined rRNA reference indices using ```bowtie``` to define the level of rRNA contamination; exports resulted statistics to file 8. Calculates isoform expression level for the sorted BAM file and GTF/TAB annotation file using GEEP reads-counting utility; exports results to file |
Path: workflows/trim-quantseq-mrnaseq-se.cwl Branch/Commit ID: 104059e07a2964673e21d371763e33c0afeb2d03 |
|
|
|
workflow_mock_ngtax.cwl
|
Path: cwl/workflows/workflow_mock_ngtax.cwl Branch/Commit ID: cd0c19d51068c5407cd70b718a561d4662819d87 |
|
|
|
Motif Finding with HOMER with custom background regions
Motif Finding with HOMER with custom background regions --------------------------------------------------- HOMER contains a novel motif discovery algorithm that was designed for regulatory element analysis in genomics applications (DNA only, no protein). It is a differential motif discovery algorithm, which means that it takes two sets of sequences and tries to identify the regulatory elements that are specifically enriched in on set relative to the other. It uses ZOOPS scoring (zero or one occurrence per sequence) coupled with the hypergeometric enrichment calculations (or binomial) to determine motif enrichment. HOMER also tries its best to account for sequenced bias in the dataset. It was designed with ChIP-Seq and promoter analysis in mind, but can be applied to pretty much any nucleic acids motif finding problem. For more information please refer to: ------------------------------------- [Official documentation](http://homer.ucsd.edu/homer/motif/) |
Path: workflows/homer-motif-analysis-bg.cwl Branch/Commit ID: 564156a9e1cc7c3679a926c479ba3ae133b1bfd4 |
|
|
|
CLIP-Seq pipeline for single-read experiment NNNNG
Cross-Linking ImmunoPrecipitation ================================= `CLIP` (`cross-linking immunoprecipitation`) is a method used in molecular biology that combines UV cross-linking with immunoprecipitation in order to analyse protein interactions with RNA or to precisely locate RNA modifications (e.g. m6A). (Uhl|Houwaart|Corrado|Wright|Backofen|2017)(Ule|Jensen|Ruggiu|Mele|2003)(Sugimoto|König|Hussain|Zupan|2012)(Zhang|Darnell|2011) (Ke| Alemu| Mertens| Gantman|2015) CLIP-based techniques can be used to map RNA binding protein binding sites or RNA modification sites (Ke| Alemu| Mertens| Gantman|2015)(Ke| Pandya-Jones| Saito| Fak|2017) of interest on a genome-wide scale, thereby increasing the understanding of post-transcriptional regulatory networks. The identification of sites where RNA-binding proteins (RNABPs) interact with target RNAs opens the door to understanding the vast complexity of RNA regulation. UV cross-linking and immunoprecipitation (CLIP) is a transformative technology in which RNAs purified from _in vivo_ cross-linked RNA-protein complexes are sequenced to reveal footprints of RNABP:RNA contacts. CLIP combined with high-throughput sequencing (HITS-CLIP) is a generalizable strategy to produce transcriptome-wide maps of RNA binding with higher accuracy and resolution than standard RNA immunoprecipitation (RIP) profiling or purely computational approaches. The application of CLIP to Argonaute proteins has expanded the utility of this approach to mapping binding sites for microRNAs and other small regulatory RNAs. Finally, recent advances in data analysis take advantage of cross-link–induced mutation sites (CIMS) to refine RNA-binding maps to single-nucleotide resolution. Once IP conditions are established, HITS-CLIP takes ~8 d to prepare RNA for sequencing. Established pipelines for data analysis, including those for CIMS, take 3–4 d. Workflow -------- CLIP begins with the in-vivo cross-linking of RNA-protein complexes using ultraviolet light (UV). Upon UV exposure, covalent bonds are formed between proteins and nucleic acids that are in close proximity. (Darnell|2012) The cross-linked cells are then lysed, and the protein of interest is isolated via immunoprecipitation. In order to allow for sequence specific priming of reverse transcription, RNA adapters are ligated to the 3' ends, while radiolabeled phosphates are transferred to the 5' ends of the RNA fragments. The RNA-protein complexes are then separated from free RNA using gel electrophoresis and membrane transfer. Proteinase K digestion is then performed in order to remove protein from the RNA-protein complexes. This step leaves a peptide at the cross-link site, allowing for the identification of the cross-linked nucleotide. (König| McGlincy| Ule|2012) After ligating RNA linkers to the RNA 5' ends, cDNA is synthesized via RT-PCR. High-throughput sequencing is then used to generate reads containing distinct barcodes that identify the last cDNA nucleotide. Interaction sites can be identified by mapping the reads back to the transcriptome. |
Path: workflows/clipseq-se.cwl Branch/Commit ID: 104059e07a2964673e21d371763e33c0afeb2d03 |
|
|
|
trim-rnaseq-pe-dutp.cwl
Runs RNA-Seq BioWardrobe basic analysis with strand specific pair-end data file. |
Path: workflows/trim-rnaseq-pe-dutp.cwl Branch/Commit ID: 12edfc2207507e53c6b5bb21e50decb5535a12f7 |
|
|
|
step-valuefrom3-wf.cwl
|
Path: cwltool/schemas/v1.0/v1.0/step-valuefrom3-wf.cwl Branch/Commit ID: fec7a10466a26e376b14181a88734983cfb1b8cb |
|
|
|
Trim Galore RNA-Seq pipeline paired-end
The original [BioWardrobe's](https://biowardrobe.com) [PubMed ID:26248465](https://www.ncbi.nlm.nih.gov/pubmed/26248465) **RNA-Seq** basic analysis for a **pair-end** experiment. A corresponded input [FASTQ](http://maq.sourceforge.net/fastq.shtml) file has to be provided. Current workflow should be used only with the single-end RNA-Seq data. It performs the following steps: 1. Trim adapters from input FASTQ files 2. Use STAR to align reads from input FASTQ files according to the predefined reference indices; generate unsorted BAM file and alignment statistics file 3. Use fastx_quality_stats to analyze input FASTQ files and generate quality statistics files 4. Use samtools sort to generate coordinate sorted BAM(+BAI) file pair from the unsorted BAM file obtained on the step 1 (after running STAR) 5. Generate BigWig file on the base of sorted BAM file 6. Map input FASTQ files to predefined rRNA reference indices using Bowtie to define the level of rRNA contamination; export resulted statistics to file 7. Calculate isoform expression level for the sorted BAM file and GTF/TAB annotation file using GEEP reads-counting utility; export results to file |
Path: workflows/trim-rnaseq-pe.cwl Branch/Commit ID: e45ab1b9ac5c9b99fdf7b3b1be396dc42c2c9620 |
|
|
|
tt_kmer_top_n.cwl
|
Path: task_types/tt_kmer_top_n.cwl Branch/Commit ID: c18a7e5164cb6b19f06b3d1e869407c118a87f7e |




